Fuellen der Luecken
1 from scipy import *
2 from pylab import *
3 from scipy.ndimage import *
4
5 def interpolate_gaps(Z):
6 ind_nan=isnan(Z) # Index of NaNs
7 sy,sx=Z.shape[0],z.shape[1]
8 Z0=Z.copy()
9 ZY=Z.copy()
10 ZX=Z.copy()
11
12 xi=arange(sx)
13 for y in range(sy):
14 L=Z[y,:]
15 ind=isfinite(L)
16 X=xi[ind]
17 Y=L[ind]
18 tck=interpolate.splrep(X,Y,s=0.0) # Calculate coefficients at the supporting points
19 spl_int=interpolate.splev(xi,tck)
20 ZY[y,:]=spl_int
21
22 Z0[ind_nan]=ZY[ind_nan]
23 return Z0
24
25
26 z=rand(60,60)*10
27 z=gaussian_filter(z,sigma=3)
28 close('all')
29 figure(1)
30 z0=z.copy()# Original image
31 z1=z.copy()# Original with missing values (set to zero)
32 #z2 temporary dilated image
33 z3=z.copy()# Reconstructed image with filled values
34
35 subplot(221)
36 imshow(z,vmin=4,vmax=6,interpolation='nearest')
37 colorbar()
38 title('Original')
39
40 # Generate missing elements set to zero
41 for i in range(30):
42 s=int(rand(1)*10)
43 ry=int(rand(1)*z.shape[0])
44 rx=int(rand(1)*z.shape[1])
45 z1[ry:ry+s,rx:rx+s]=nan
46
47 ind_zero=(z1==0).nonzero() # Index of elements that are zero
48
49 z2=z1.copy()
50 z3=interpolate_gaps(z2)
51
52
53 subplot(222)
54 imshow(z1,vmin=4,vmax=6,interpolation='nearest')
55 colorbar()
56 title('Missing data')
57
58 subplot(223)
59 imshow(z3,vmin=4,vmax=6,interpolation='nearest')
60 colorbar()
61 title('Filled gaps')
62
63 subplot(224)
64 imshow(z0-z3,interpolation='nearest')
65 colorbar()
66 title('Difference')
67 savefig('filling_gaps.png',dpi=50)
68 show()
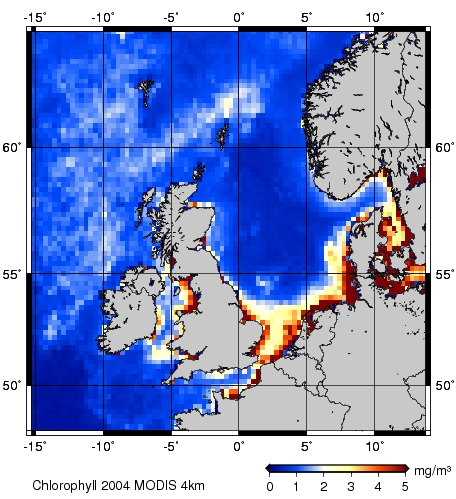
Erzeugung und Speicherung einer Testdatei für die gmt-Darstellung
Eine Erklärung zu diesem Programm findet man unter ChlorophyllArbeitsGruppe2 .
1 from scipy import *
2 import struct
3 from pyhdf.SD import *
4 from pylab import *
5
6 def read_CHLO(filename):
7 # Oeffne HDF4-Datei
8 f=SD(filename)
9 # Hole die Attribute (Metadata)
10 attr = f.attributes()
11 # Hole die in der Datei verfuegbaren Datensaetze
12 dsets = f.datasets()
13 # Hole die Daten aus der Datei in ein scipy.array
14 data=array(f.select('l3m_data').get())
15 # Siehe attr:
16 #'Scaling Equation': ('Base**((Slope*l3m_data) + Intercept) = Parameter value\x00',
17 Base=attr['Base']
18 Slope=attr['Slope']
19 Intercept=attr['Intercept']
20 data[(data==65535).nonzero()]=nan
21 # Skaliere die Daten, um die urspruenglichen Einheiten zu berechnen
22 data=Base**((Slope*data) + Intercept)
23 return data,attr
24
25 def hdf_set(filename,z):
26 hdfFile=filename
27 f = SD(hdfFile,SDC.WRITE|SDC.CREATE)
28 f.author = 'Lars Kaleschke'
29 v2=f.create('z',SDC.FLOAT32,(z.shape[0],z.shape[1]))
30 v2[:]=z.astype(float32)
31 f.end()
32 return
33
34
35 filename='/pf/u/u243066/satdat/chlorophyll/Modis_2004_YR_chlo_9'
36 img,metadata=read_CHLO(filename)
37
38 ##ECOHAM3/4-Gebiet ausschneiden
39 ##ECOHAM3/4 Latitude:47.5833 to 63.9833, Longitude:-15.25 to 14.0833
40
41 ecolat1=63.9833
42 ecolat2=47.5833
43 ecolon1=-15.25
44 ecolon2=14.0833
45 latstep=float(metadata['Latitude Step'])
46 lonstep=float(metadata['Longitude Step'])
47 Nlat=float(metadata['Northernmost Latitude'])
48 Wlon=float(metadata['Westernmost Longitude'])
49
50 lat1=floor((Nlat-ecolat1)/latstep)
51 lat2=ceil((Nlat-ecolat2)/latstep)
52 lon1=floor((abs(Wlon-ecolon1))/lonstep)
53 lon2=ceil((abs(Wlon-ecolon2))/lonstep)
54 img_eco34=img[lat1:lat2,lon1:lon2]
55
56 #Anpassung des Satellitenbildausschnittes auf Modellgitter:
57 img_eco34_modgit=ndimage.zoom(img_eco34,(82./198.,88./353.))
58
59 #Speichern des angepassten Satbildes als hdf-Datei:
60
61 filename='testbild82x88.hdf'
62 hdf_set(filename,img_eco34_modgit)
gmt-Darstellung
1 from scipy import *
2 import struct,pipes,os
3 from pyhdf.SD import *
4
5 def hdf_get(filename):
6 f = SD(filename)
7 dsets = f.datasets()
8 d={}
9 for dset in dsets.keys():
10 ds=f.select(dset)
11 d[dset]=array(ds.get(),float)
12 return d
13
14
15 def write_grd(data,lat,lon,filename,I,R):
16
17 # Write to GMT/NetCDF .grd
18
19 grdtable,grdfile=filename+'.tab',filename
20 pipe=pipes.Template()
21 cmd='xyz2grd -V -bis '+I+' '+R+' -G'+grdfile
22 print cmd
23 pipe.append(cmd,'-.')
24 f=pipe.open(grdtable, 'w')
25 sy,sx=data.shape[0],data.shape[1]
26 for yi in range(0,sy):
27 for xi in range(0,sx):
28 f.write(struct.pack("f",lon[yi,xi])+struct.pack("f",lat[yi,xi])+struct.pack("f",data[yi,xi]))
29 f.close()
30
31 return
32
33
34 def gmt_map(data,outfile,title):
35 Dlon=1.0/3
36 Dlat=0.2
37
38 X=88
39 Y=82
40
41 upper_lat=63.983
42 lower_lat=upper_lat-Dlat*Y+Dlat
43 western_lon=-15.25
44 eastern_lon=western_lon+Dlon*X
45
46 R=' -R%f'%western_lon+'/%f'%eastern_lon+'/%f'%lower_lat+'/%f'%upper_lat
47 I=' -I0.3333333/0.2'
48 J=' -Jm0.2 '
49 A=' -Ba5g5/a5g5NESW '
50 C='color.tab'
51
52 lats=(upper_lat-indices((Y,X))[0,:,:].astype(float)*Dlat)
53 lons=(indices((Y,X))[1,:,:].astype(float)*Dlon+western_lon)
54
55 filename='data.grd'
56 write_grd(data,lats,lons,filename,I,R)
57
58
59 os.system('gmtset PAPER_MEDIA A4 PLOT_DEGREE_FORMAT ddd:mm:ss')
60 os.system('makecpt -Cjet -T0/5/1 -Z -D > '+C)
61 os.system('grdimage '+filename+R+J+A+' -C'+C+' -K > '+outfile)
62 os.system('pscoast '+R+J+A+' -N1 -W1/0/0/0 -G200/200/200 -Di -K -O >> '+outfile)
63
64 #Scale bar
65 scl_tick='1/:mg/m\032:'
66 print 'Add scale bar'
67 os.system('psscale -D3.5/-0.6/2.0/0.1h -E -C'+C+' -B'+scl_tick+' -K -O >> '+outfile)
68 os.system('echo "0.0 -0.03 14 0.0 0 0 '+title+'" | pstext -N -R0/1/0/1 -JX21/29 -O >> '+outfile)
69 return
70
71
72
73 #TEST
74 filename='testbild82x88.hdf'
75 d=hdf_get(filename)
76
77 data=d['z']
78 outfile='map.ps'
79 title='Chlorophyll am Tag X'
80 gmt_map(data,outfile,title)
81 os.system('gv '+outfile)
GMT Karte