Contents
Input/output and data formats
This lesson deals with the ways of reading and writing data in different formats
Basic Python
The file object can be used for reading and writing plain text as well as unformatted binary data. The following code writes a message in the file with the name out.txt, reads and print the data
write() writes a string to the file
read() reads complete file
read(N) reads N bytes
readlines() reads the file with linebreaks
readline() reads only the next line
Pickle
The pickle module implements an algorithm for serializing and de-serializing a Python object structure. Pickling is the process whereby a Python object hierarchy is converted into a byte stream, and unpickling is the inverse operation, whereby a byte stream is converted back into an object hierarchy. The cPickle module is a much faster implementation and should be preferred.
Comma Separated Values
The so-called CSV (Comma Separated Values) format is the most common import and export format for spreadsheets (Excel) and databases. The csv module enables CSV file reading and writing
NumPy/SciPy
HDF
NASA's standard file format, the http://hdf.ncsa.uiuc.edu/index.html is a self-describing data format. HDF files can contain binary data and allow direct access to parts of the file without first parsing the entire contents.
The HDF versions 4 and 5 are not compatible.
Different modules are available for reading and writing HDF files
pyhdf
pyhdf is a python interface to the NCSA HDF4 library.
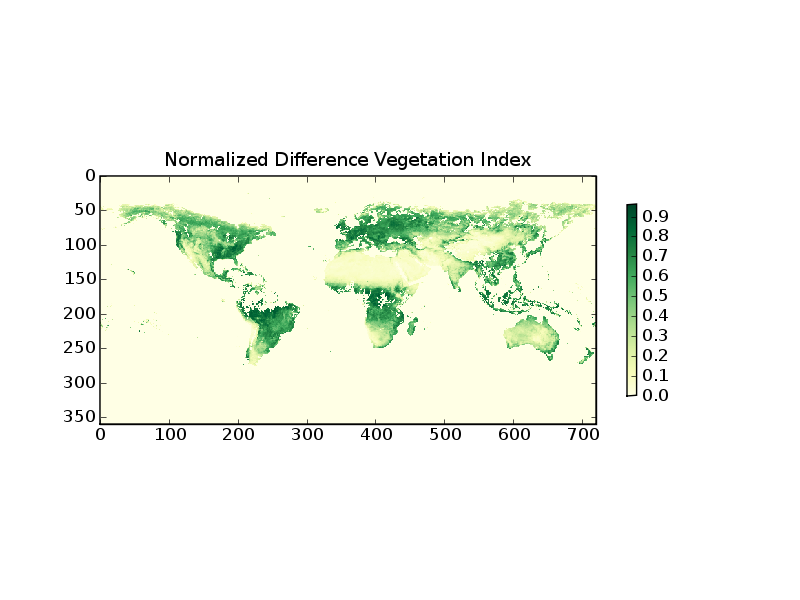
The following example demonstrates how to read level-3 data from the Multi-angle Imaging Spectral Radiometer (MISR) on the Terra Satellite
1 from scipy import array
2 from pylab import imshow,colorbar,title,savefig
3 from pyhdf.SD import SD
4
5 f=SD('MISR_AM1_CGLS_MAY_2007_F04_0025.hdf')
6 print f.datasets().keys()
7 data=array(f.select('NDVI average').get())
8 data[data<0]=0
9
10 imshow(data,interpolation='nearest',cmap=cm.YlGn)
11 colorbar(shrink=0.5)
12 title('Normalized Difference Vegetation Index')
Line 5 opens the HDF file object. line 6 prints the keywords of the included datasets. From this one can identify the keyword for the desired parameter. Line 7 reads the data in a SciPy array. Line 8 selects the negative (bad and missing) data and sets them to zero.
netCDF
