|
Size: 3394
Comment:
|
← Revision 15 as of 2011-01-17 13:34:14 ⇥
Size: 4057
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| #acl AdminGroup:read,write,revert,delete EditorGroup:read,write,revert,delete StudentGroup:read,write | #acl AdminGroup:read,write,revert,delete EditorGroup:read,write,revert,delete StudentGroup:read,write All:read |
| Line 13: | Line 13: |
| == Example: ARGO floats == | = Example data from Argo system = |
| Line 15: | Line 15: |
| http://en.wikipedia.org/wiki/Argo_%28oceanography%29 | [[http://en.wikipedia.org/wiki/Argo_%28oceanography%29|Argo observation system]] |
| Line 19: | Line 19: |
| First we have to download the data. Here is a [[/download script]] that can be used to retrieve one month of data in a temporary directory called {{{tmp_dir}}}: | First we have to download the data. Here is a [[/download script]] that can be used to retrieve one month of Argo data. |
| Line 21: | Line 21: |
| {{{#!python #!/usr/bin/env python import os,os.path from ftplib import FTP |
We extract only the surface temperature to reduce the large amount of data. This [[/extract script]] generates an overview of the data and writes the surface temperature in a file [[attachment:lat_lon_T.tab]] which contains latitude, longitude and surface temperature. We can use this file in the following. |
| Line 26: | Line 23: |
| tmp_dir='/scratch/clisap/seaice/TMP/u242023/ARGO/' | |
| Line 28: | Line 24: |
| ftp_adr='ftp.ifremer.fr' ftp_dir='/ifremer/argo/geo/atlantic_ocean/2011/01/' |
{{attachment:argo_position.png}} |
| Line 31: | Line 26: |
| ftp = FTP(ftp_adr) # connect to host, default port ftp.login() # user anonymous, passwd anonymous@ ftp.cwd(ftp_dir) |
Argo positions of measurements |
| Line 35: | Line 28: |
| file_list=ftp.nlst() | {{attachment:Argo_plot.png}} |
| Line 37: | Line 30: |
| for f in file_list: urlfile='ftp://'+ftp_adr+ftp_dir+f localfile=tmp_dir+f if not(os.path.exists(localfile)): print 'Getting file '+f #We use curl instead of ftp.retrbinary for download os.system("curl -s -k -o "+localfile+" "+urlfile) else: print 'file '+f+' exists' |
Measured temperature profiles (unfiltered data). |
| Line 48: | Line 32: |
| ftp.close() | = Tools for 2-dimensional interpolation and gridding = == Python == {{{ Help on function griddata in module matplotlib.mlab: griddata(x, y, z, xi, yi) ``zi = griddata(x,y,z,xi,yi)`` fits a surface of the form *z* = *f*(*x*, *y*) to the data in the (usually) nonuniformly spaced vectors (*x*, *y*, *z*). :func:`griddata` interpolates this surface at the points specified by (*xi*, *yi*) to produce *zi*. *xi* and *yi* must describe a regular grid, can be either 1D or 2D, but must be monotonically increasing. A masked array is returned if any grid points are outside convex hull defined by input data (no extrapolation is done). Uses natural neighbor interpolation based on Delaunay triangulation... |
| Line 51: | Line 54: |
| The next script generates an overview of the data and writes the surface temperature in a file called {{{lat_lon_T.tab}}} which contains latitude, longitude and surface temperature. [[attachment:lat_lon_T.tab]] | == GMT == |
| Line 53: | Line 56: |
| {{attachment:argo_position.png}} {{attachment:Argo_plot.png}} {{{#!python import scipy.io as io import glob from pylab import * from mpl_toolkits.basemap import Basemap tmp_dir='/scratch/clisap/seaice/TMP/u242023/ARGO/' |
[[http://gmt.soest.hawaii.edu/|GMT]] |
| Line 66: | Line 59: |
| file_liste=glob.glob(tmp_dir+'*.nc') D={}# Empty dictionary to store selected profiles for f in file_liste:# Loop over all data |
[[http://gmt.soest.hawaii.edu/gmt/doc/gmt/html/GMT_Tutorial/node44.html|Nearest neighbor gridding]] with 5 degree grid cell size {{{ nearneighbor lat_lon_T.tab -Rd -I300m -S300m -N1 -Ggrid.nc }}} Generate color table: {{{ makecpt -Crainbow -T-2/30/1 > g.cpt }}} Plot grid files in 2-D {{{ grdimage grid.nc -Rd -JG-45/0/4i -Cg.cpt -K > out.ps }}} Plot continents on map {{{ pscoast -Rd -JG-45/0/4i -B15g15 -Dc -Gblack -O >> out.ps }}} Common options: {{{ -G name of output grid. -R specifies the min/max coordinates of data region in user units. -J Selects map projection. -K Means allow for more plot code to be appended later. -O means Overlay plot mode. }}} |
| Line 71: | Line 89: |
| # Open netcdf data file fid=io.netcdf_file(f,'r') |
Show resulting postscript map {{{ gv out.ps }}} |
| Line 74: | Line 94: |
| # Read content into variables lat=fid.variables['LATITUDE'][:].copy() lon=fid.variables['LONGITUDE'][:].copy() T=fid.variables['TEMP'][:].copy() P=fid.variables['PRES'][:].copy() T[T>=99999]=nan # Set 99999.0 to "Not a Number" P[P>=99999]=nan |
Convert to png format for this Wiki page {{{ convert -trim out.ps out.png }}} |
| Line 83: | Line 99: |
| (nr_profs,Z)=T.shape # Get dimension for i in range(nr_profs): D[(lon[i],lat[i])]=(T[i,:],P[i,:]) # Close data file fid.close() fid=open('lat_lon_T.tab','w') for k in D.keys():# write position (lat,lon), surface temperature to file fid.write(str(k[0])+'\t'+str(k[1])+'\t'+str(D[k][0][0])+'\n') fid.close() stop |
Convert postscript to pdf format (you can zoom in the map with acroread) {{{ ps2pdf out.ps acroread out.pdf }}} |
| Line 98: | Line 106: |
| # Draw map of positions m = Basemap(projection='ortho',lon_0=-45,lat_0=0,resolution='l') m.bluemarble() for lon,lat in D.keys(): x,y=m(lon,lat) # Coordinate transfer m.plot(x,y,'r.') savefig('argo_position.png',dpi=150) |
|
| Line 107: | Line 107: |
| # Plot profiles figure() for k in D.keys(): # print k plot(D[k][0][:],D[k][1][:]) axis([-2,30,2000,0]) xlabel('T') ylabel('P') show() savefig('Argo_plot.png',dpi=75) }}} |
{{attachment:out.png}} === Exercises === Read the GMT manual: * Find out the meaning of the options I, S, N of the nearest neighbor gridding * What does -Rd mean? * What does -Dc mean? * What projection was chosen? Produce new plots: * Select a sub region of the data, i.e. only the North Atlantic. * Select a different projection * Plot the data with this projection Use new interpolation parameters: * Modify grid cell size (increment) to obtain a finer grid * Modify search radius Interaction with Python: * Read the GMT-netcdf file into Python with {{{scipy.io.netcdf_file}}}, (variable z) * Write a Python script that does the interpolation with GMT by using {{{os.system()}}} Use different interpolation technique: * {{{surface}}}, adjustable tension continuous curvature surface gridding algorithm |
Contents
2-dimensional interpolation and gridding
2-dimensional interpolation and gridding is a common problem for the representation of measurements on a map. Usually measurements are taken at irregular sample points and not in a regular grid. There are various approaches for the problem and the best solution depends on the data.
In the following we will look at oceanographic parameters that have been measured with the Argo system.
Example data from Argo system
Data are provided at, i.e. ftp://ftp.ifremer.fr//ifremer/argo/
First we have to download the data. Here is a /download script that can be used to retrieve one month of Argo data.
We extract only the surface temperature to reduce the large amount of data. This /extract script generates an overview of the data and writes the surface temperature in a file lat_lon_T.tab which contains latitude, longitude and surface temperature. We can use this file in the following.
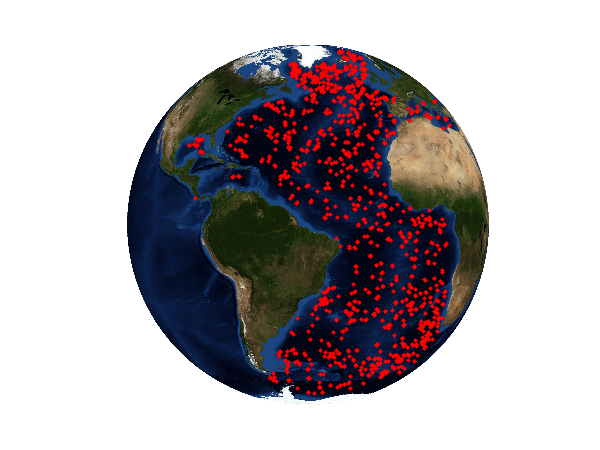
Argo positions of measurements
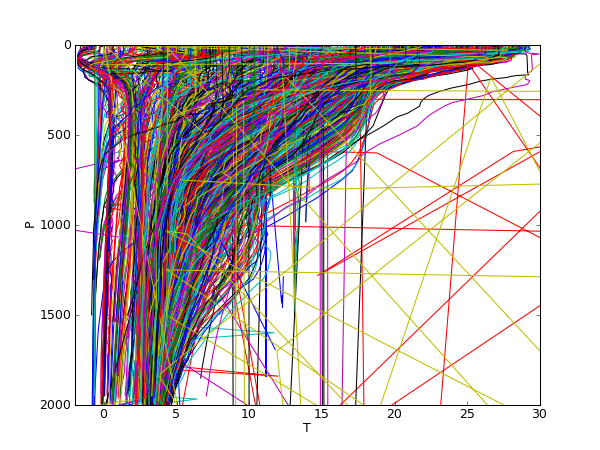
Measured temperature profiles (unfiltered data).
Tools for 2-dimensional interpolation and gridding
Python
Help on function griddata in module matplotlib.mlab:
griddata(x, y, z, xi, yi)
``zi = griddata(x,y,z,xi,yi)`` fits a surface of the form *z* =
*f*(*x*, *y*) to the data in the (usually) nonuniformly spaced
vectors (*x*, *y*, *z*). :func:`griddata` interpolates this
surface at the points specified by (*xi*, *yi*) to produce
*zi*. *xi* and *yi* must describe a regular grid, can be either 1D
or 2D, but must be monotonically increasing.
A masked array is returned if any grid points are outside convex
hull defined by input data (no extrapolation is done).
Uses natural neighbor interpolation based on Delaunay
triangulation...
GMT
Nearest neighbor gridding with 5 degree grid cell size
nearneighbor lat_lon_T.tab -Rd -I300m -S300m -N1 -Ggrid.nc
Generate color table:
makecpt -Crainbow -T-2/30/1 > g.cpt
Plot grid files in 2-D
grdimage grid.nc -Rd -JG-45/0/4i -Cg.cpt -K > out.ps
Plot continents on map
pscoast -Rd -JG-45/0/4i -B15g15 -Dc -Gblack -O >> out.ps
Common options:
-G name of output grid. -R specifies the min/max coordinates of data region in user units. -J Selects map projection. -K Means allow for more plot code to be appended later. -O means Overlay plot mode.
Show resulting postscript map
gv out.ps
Convert to png format for this Wiki page
convert -trim out.ps out.png
Convert postscript to pdf format (you can zoom in the map with acroread)
ps2pdf out.ps acroread out.pdf
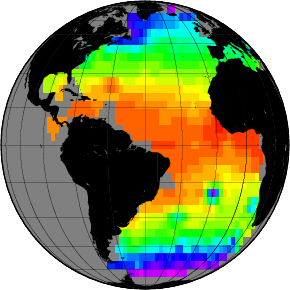
Exercises
Read the GMT manual:
- Find out the meaning of the options I, S, N of the nearest neighbor gridding
- What does -Rd mean?
- What does -Dc mean?
- What projection was chosen?
Produce new plots:
- Select a sub region of the data, i.e. only the North Atlantic.
- Select a different projection
- Plot the data with this projection
Use new interpolation parameters:
- Modify grid cell size (increment) to obtain a finer grid
- Modify search radius
Interaction with Python:
Read the GMT-netcdf file into Python with scipy.io.netcdf_file, (variable z)
Write a Python script that does the interpolation with GMT by using os.system()
Use different interpolation technique:
surface, adjustable tension continuous curvature surface gridding algorithm
